Complementary DNA/RNA-Based Profiling: Characterization of Corrosive Microbial Communities and Their Functional Profiles in an Oil Production Facility

Overview
Corrosion in oil production facilities represents a chief concern to the energy sector because it compromises the integrity of the metallic infrastructure, resulting in equipment failure before their expected lifetime and severe environmental damage. Microorganisms can cause or promote corrosion by their interaction with the metallic surface, a phenomenon that drives a worldwide market for microbial control that is worth billions of dollars annually. However, identification of mechanisms and implicated microbial populations remains a challenge due to the plethora of microbes typically identified using DNA-based surveys which provide little information about the active members of the microbial community, likely to be the ones contributing to corrosion. In this paper, it is demonstrated that a complementary approach using RNA-based profiling can elucidate the active populations in the corrosive environment which helps identify the corrosion mechanism and the metabolic pathways possibly involved in the corrosion reactions. The comparison of the total versus active microbial populations through several bioinformatics tools in this research, provided valuable information about how microbial structure is affected by the operational conditions in the oilfield and revealed the persistent populations that remain active upon biocide treatment thus providing insights into the efficacy of treatments to control microbiologically influenced corrosion.
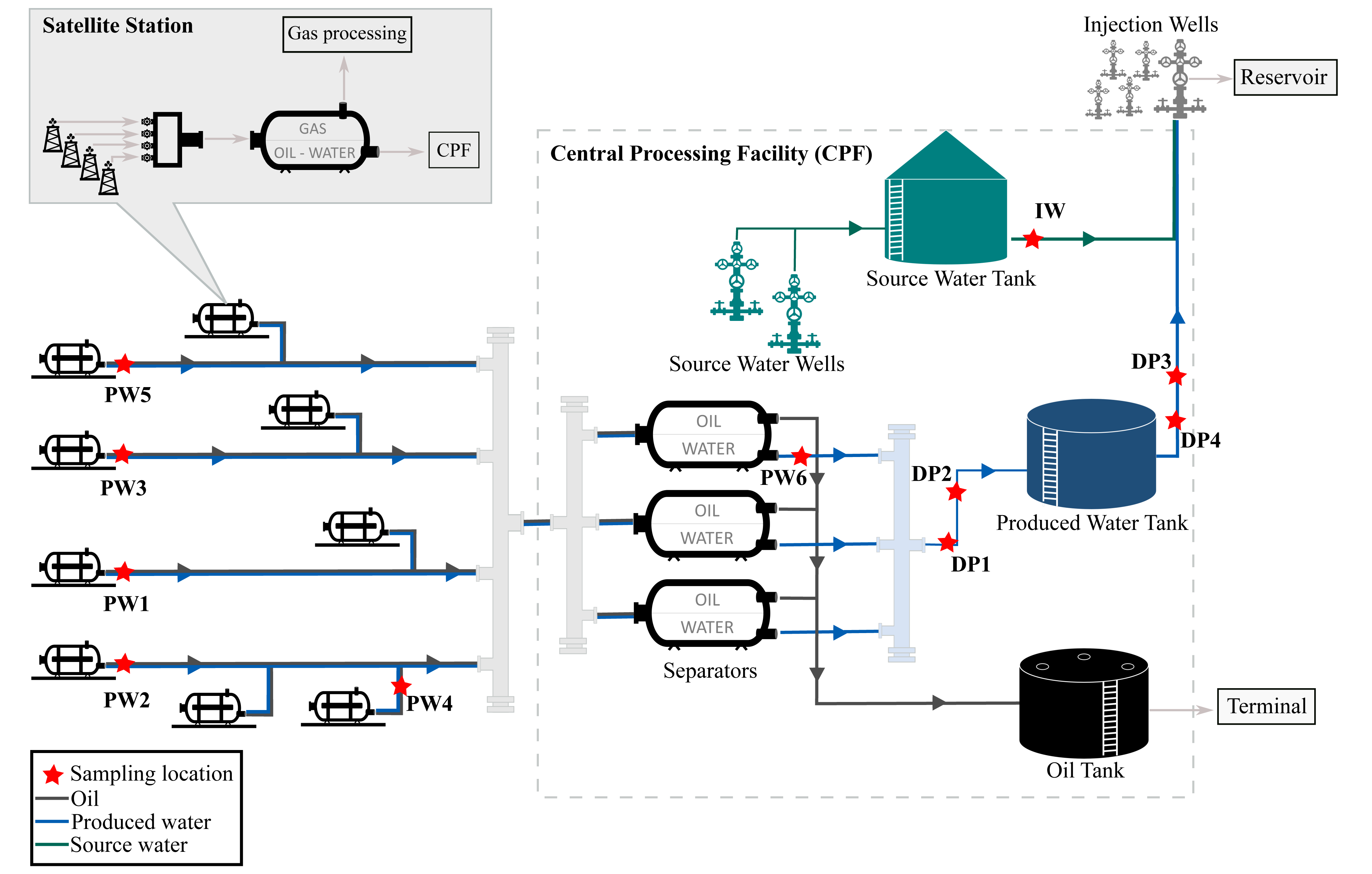
Figure 1; Process flow diagram of the oil production facility.
Objectives
This work aimed to determine whether the application of complementary analysis using amplicon sequencing of the 16S rRNA gene and transcripts would provide relevant information on the microbial communities recovered from an oil production facility with corrosion issues.
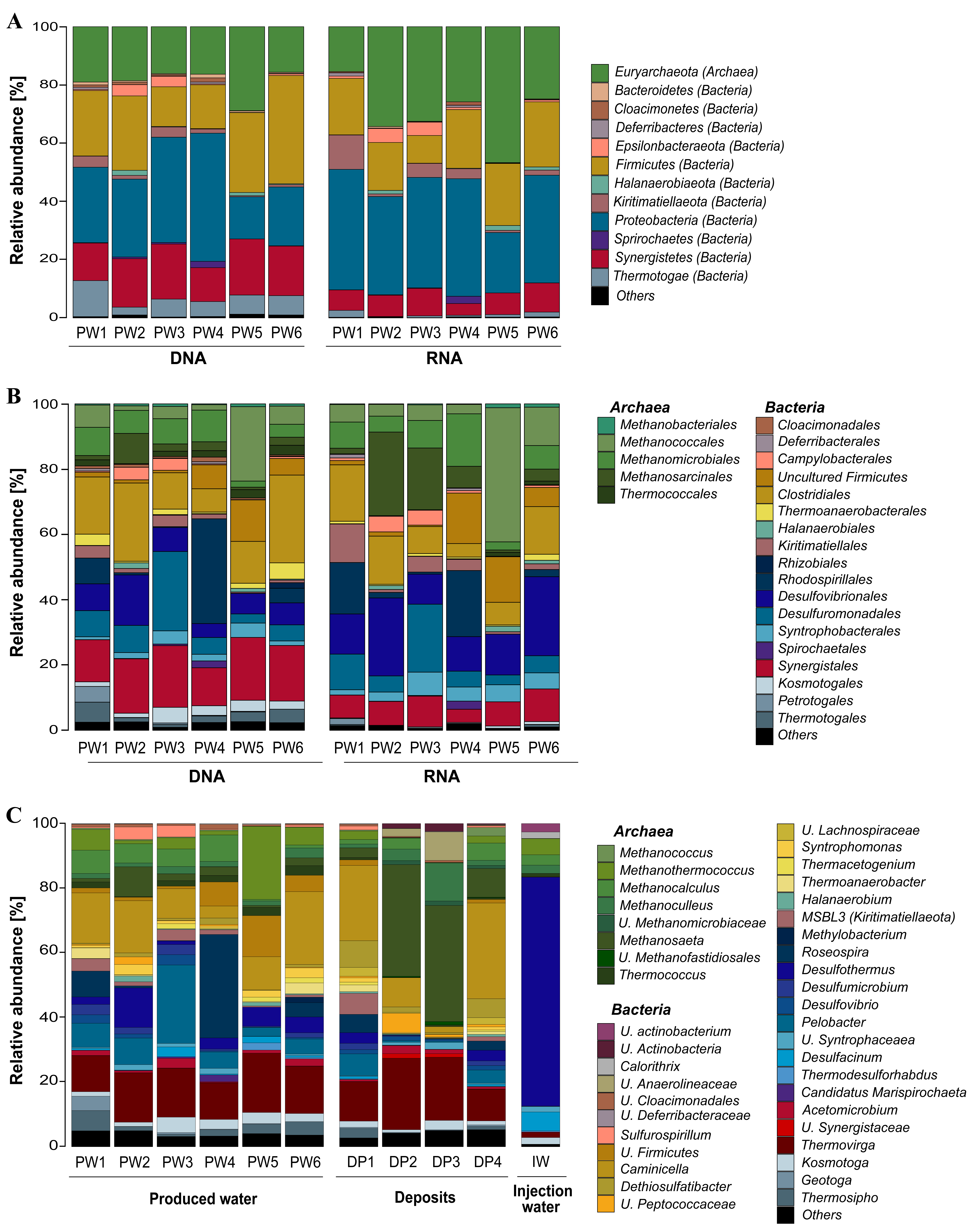
Figure 2; Microbial community composition in the oilfield retrieved from 16S rRNA gene and transcripts sequencing. (A) Total and active microbial communities in produced water samples at phylum level. (B) Total and active microbial communities in produced water samples at order level. (C) Total microbial community in produced water, deposits and injection water samples at genus level. U., unclassified. Phylogenetic groups accounting for <1% of all classified sequences were summarized in the artificial group “Others.” Results from the three replicates collected in each sampling location were pooled together.
Why is it important?
Microbiologically influenced corrosion (MIC) management of industry assets involves a microbiological assessment to detect the presence of MIC causative microorganisms. In this investigation, it was showed that DNA-based sequencing results alone could lead to underestimation of active members in the community resulting in an inaccurate MIC risk assessment. We demonstrated that a complementary methodological analysis using DNA and RNA-based sequencing provides a better representation of the total and active communities in industrial facilities experiencing corrosion, information that can improve the understanding, mitigation and prediction of MIC processes.
Get the paper!
Citation
S.J. Salgar-Chaparro, L.L. Machuca, Front Microbiol 10 (2019): p. 2587.